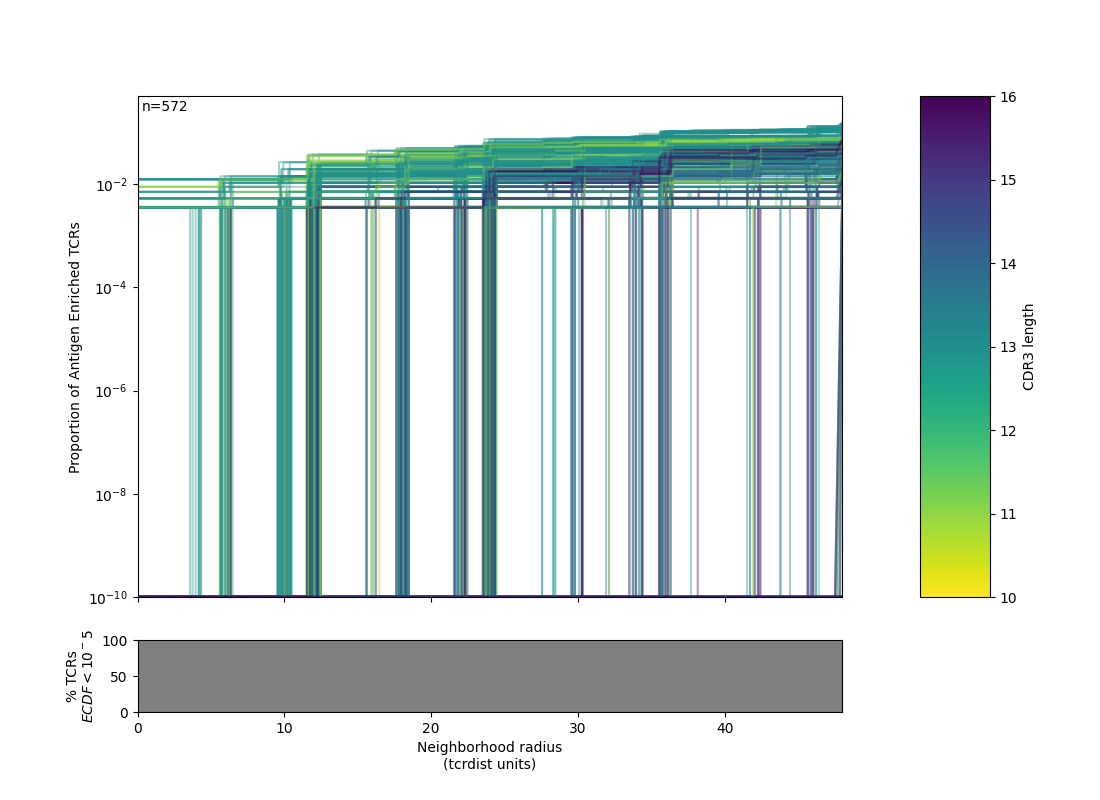
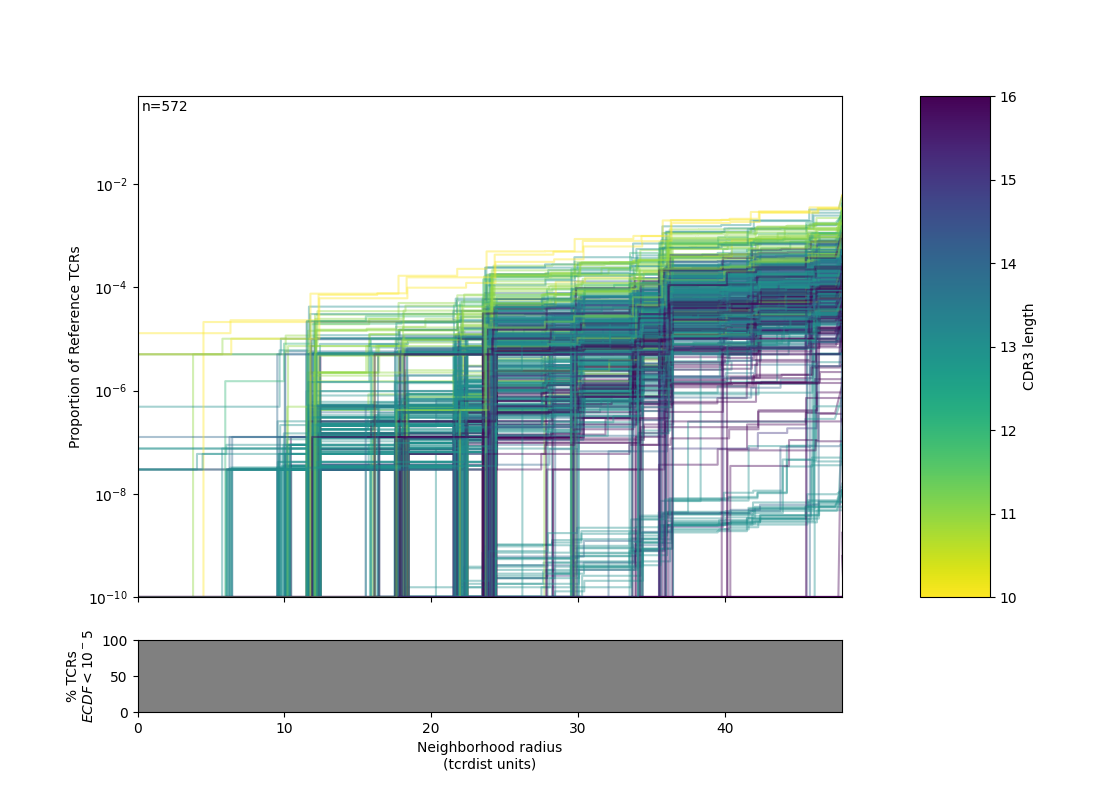
Meta-Clonotypes¶
The script for meta-clonotype discovery contains a function (find_metaclonotypes). It encapsulates a workflow we used for finding meta-clonotypes in antigen-enriched data as
described in ‘TCR meta-clonotypes for biomarker discovery with tcrdist3: quantification of public, HLA-restricted TCR biomarkers of SARS-CoV-2 infection’
bioRxiv 2020 .
To ensure you have the necessary background reference files in your environment or the tcrdist3 docker container, run the following before getting started:
apt-get install unzip
python3 -c "from tcrsampler.setup_db import install_all_next_gen; install_all_next_gen(dry_run = False)"
Meta-Clonotype Discovery¶
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 73 74 75 76 77 78 79 80 81 82 83 84 85 86 87 88 89 90 91 92 93 94 95 96 97 98 99 100 101 102 103 104 105 106 107 108 109 110 111 112 113 114 115 116 117 118 119 120 121 122 123 124 125 126 127 128 129 130 131 132 133 134 135 136 137 138 139 140 141 142 143 144 145 146 147 148 149 150 151 152 153 154 155 156 157 158 159 160 161 162 163 164 165 166 167 168 169 170 171 172 173 174 175 176 177 178 179 180 181 182 183 184 185 186 187 188 189 190 191 192 193 194 195 196 197 198 199 200 201 202 203 204 205 206 207 208 209 210 211 212 213 214 215 216 217 218 219 220 221 222 223 224 225 226 227 228 229 230 231 232 233 234 235 236 237 238 239 240 241 242 243 244 245 246 247 248 249 250 251 252 253 254 255 256 257 258 259 260 261 262 263 264 265 266 267 268 269 270 271 272 273 274 275 276 277 278 279 280 281 282 283 284 285 286 287 288 289 290 291 292 293 294 295 296 297 298 299 300 301 302 303 304 305 306 307 308 309 310 311 312 313 314 315 316 317 318 319 320 321 322 323 324 325 326 327 328 329 330 331 332 333 334 335 336 337 338 339 340 341 342 343 344 345 346 347 348 349 350 351 352 353 354 355 356 357 358 359 360 361 362 363 364 365 366 367 368 369 370 371 372 373 374 375 376 377 378 379 380 381 382 383 384 385 386 387 388 389 390 391 392 393 394 395 396 397 398 399 400 401 402 403 404 405 406 407 408 409 410 411 412 413 414 415 416 417 418 419 420 421 422 423 424 425 426 427 428 429 430 431 432 433 434 435 436 437 438 439 440 441 442 443 444 445 446 447 448 449 450 451 452 453 454 455 456 457 458 459 460 461 462 463 464 465 466 467 468 469 470 471 472 473 474 475 476 477 478 479 480 481 482 483 484 485 486 487 488 489 490 491 492 493 494 495 496 497 498 499 500 501 502 503 504 505 506 507 508 509 510 511 512 513 514 515 516 517 518 519 520 521 522 523 524 525 526 527 | """ 2020-12-18 Seattle, WA HLA CLASS I restricted meta-clonotype discovery. This functions encapsulates a complete work flow for finding meta-clonotypes in antigen-enriched data. prepare docker container: apt-get install unzip python3 -c "from tcrsampler.setup_db import install_all_next_gen; install_all_next_gen(dry_run = False)" """ import sys import os import numpy as np import pandas as pd import scipy.sparse import matplotlib.pyplot as plt from tcrdist.paths import path_to_base from tcrdist.repertoire import TCRrep from tcrdist.automate import auto_pgen from tcrsampler.sampler import TCRsampler from tcrdist.background import get_stratified_gene_usage_frequency from tcrdist.background import sample_britanova from tcrdist.background import make_gene_usage_counter, get_gene_frequencies from tcrdist.background import calculate_adjustment, make_gene_usage_counter from tcrdist.background import make_vj_matched_background, make_flat_vj_background from tcrdist.ecdf import distance_ecdf, _plot_manuscript_ecdfs from tcrdist.centers import calc_radii from tcrdist.public import _neighbors_variable_radius from tcrdist.public import _neighbors_sparse_variable_radius from tcrdist.regex import _index_to_regex_str def find_metaclonotypes( project_path = "tutorial48", source_path = os.path.join(path_to_base,'tcrdist','data','covid19'), antigen_enriched_file = 'mira_epitope_48_610_YLQPRTFL_YLQPRTFLL_YYVGYLQPRTF.tcrdist3.csv', ncpus = 4, seed = 3434): """ This functions encapsulates a complete workflow for finding meta-clonotypes in antigen-enriched data. """ np.random.seed(seed) if not os.path.isdir(project_path): os.mkdir(project_path) ############################################################################ # Step 1: Select and load a antigen-enriched (sub)repertoire. #### ############################################################################ print(f"INITIATING A TCRrep() with {antigen_enriched_file}") assert os.path.isfile(os.path.join(source_path, antigen_enriched_file)) # Read file into a Pandas DataFrame <df> df = pd.read_csv(os.path.join(source_path, antigen_enriched_file)) # Drop cells without any gene usage information df = df[( df['v_b_gene'].notna() ) & (df['j_b_gene'].notna()) ] # Initialize a TCRrep class, using ONLY columns that are complete and unique define a a clone. # Class provides a 'count' column if non is present # Counts of identical subject:VCDR3 'clones' will be aggregated into a TCRrep.clone_df. from tcrdist.repertoire import TCRrep tr = TCRrep(cell_df = df[['subject','cell_type','v_b_gene', 'j_b_gene', 'cdr3_b_aa']], organism = "human", chains = ['beta'], compute_distances = True) tr.cpus = ncpus ############################################################################ # Step 1.1: Estimate Probability of Generation #### ############################################################################ ### It will be useful later to know the pgen of each from tcrdist.automate import auto_pgen print(f"COMPUTING PGEN WITH OLGA (Sethna et al 2018)") print("FOR ANTIGEN-ENRICHED CLONES TO BE USED FOR SUBSEQUENT ANALYSES") auto_pgen(tr) # Tip: Users of tcrdist3 should be aware that by default a <TCRrep.clone_df> # DataFrame is created out of non-redundant cells in the cell_df, and # pairwise distance matrices automatically computed. # Notice that attributes <tr.clone_df> and <tr.pw_beta> , <tr.pw_cdr3_b_aa>, # are immediately accessible. # Attributes <tr.pw_pmhc_b_aa>, <tr.pw_cdr2_b_aa>, and <tr.pw_cdr1_b_aa> # are also available if <TCRrep.store_all_cdr> is set to True. # For large datasets, i.e., >15,000 clones, this approach may consume too much # memory so <TCRrep.compute_distances> is automatically set to False. ############################################################################ # Step 2: Synthesize an Inverse Probability Weighted VJ Matched Background # ############################################################################ # Generating an appropriate set of unenriched reference TCRs is important; for # each set of antigen-associated TCRs, discovered by MIRA, we created a two part # background. One part consists of 100,000 synthetic TCRs whose V-gene and J-gene # frequencies match those in the antigen-enriched repertoire, using the software # OLGA (Sethna et al. 2019; Marcou et al. 2018). The other part consists of # 100,000 umbilical cord blood TCRs sampled uniformly from 8 subjects (Britanova # et al., 2017). This mix balances dense sampling of sequences near the # biochemical neighborhoods of interest with broad sampling of TCRs from an # antigen-naive repertoire. Importantly, we adjust for the biased sampling by # using the V- and J-gene frequencies observed in the cord-blood data (see # Methods for details about inverse probability weighting adjustment). Using this # approach we are able to estimate the abundance of TCRs similar to a centroid # TCR in an unenriched background repertoire of ~1,000,000 TCRs, using a # comparatively modest background dataset of 200,000 TCRs. While this estimate # may underestimate the true specificity, since some of the neighborhood TCRs in # the unenriched background repertoire may in fact recognize the antigen of # interest, it is useful for prioritizing neighborhoods and selecting a radius # for each neighborhood that balances sensitivity and specificity. # Initialize a TCRsampler -- human, beta, umbilical cord blood from 8 people. print(f"USING tcrsampler TO CONSTRUCT A CUSTOM V-J MATCHED BACKGROUND") from tcrsampler.sampler import TCRsampler ts = TCRsampler(default_background = 'britanova_human_beta_t_cb.tsv.sampler.tsv') # Stratify sample so that each subject contributes similarly to estimate of # gene usage frequency from tcrdist.background import get_stratified_gene_usage_frequency ts = get_stratified_gene_usage_frequency(ts = ts, replace = True) # Synthesize an inverse probability weighted V,J gene background that matches # usage in your enriched repertoire df_vj_background = tr.synthesize_vj_matched_background(ts = ts, chain = 'beta') # Get a randomly drawn stratified sampler of beta, cord blood from # Britanova et al. 2016 # Dynamics of Individual T Cell Repertoires: From Cord Blood to Centenarians from tcrdist.background import sample_britanova df_britanova_100K = sample_britanova(size = 100000) # Append frequency columns using, using sampler above df_britanova_100K = get_gene_frequencies(ts = ts, df = df_britanova_100K) df_britanova_100K['weights'] = 1 df_britanova_100K['source'] = "stratified_random" # Combine the two parts of the background into a single DataFrame df_bkgd = pd.concat([df_vj_background.copy(), df_britanova_100K.copy()], axis = 0).\ reset_index(drop = True) # Assert that the backgrounds have the expected number of rows. assert df_bkgd.shape[0] == 200000 # Save the background for future use background_outfile = os.path.join(project_path, f"{antigen_enriched_file}.olga100K_brit100K_bkgd.csv") print(f'WRITING {background_outfile}') df_bkgd.to_csv(background_outfile, index = False) # Load the background to a TCRrep without computing pairwise distances # (i.e., compute_distances = False) tr_bkgd = TCRrep( cell_df = df_bkgd, organism = "human", chains = ['beta'], compute_distances = False) # Compute rectangular distances. Those are, distances between each clone in # the antigen-enriched repertoire and each TCR in the background. # With a single 1 CPU and < 10GB RAM, 5E2x2E5 = 100 million pairwise distances, # across CDR1, CDR2, CDR2.5, and CDR3 # 1min 34s ± 0 ns per loop (mean ± std. dev. of 1 run, 1 loop each) # %timeit -r 1 tr.compute_rect_distances(df = tr.clone_df, df2 = tr_bkdg.clone_df, store = False) ############################################################################ # Step 4: Calculate Distances ##### ############################################################################ print(f"COMPUTING RECTANGULARE DISTANCE") tr.compute_sparse_rect_distances( df = tr.clone_df, df2 = tr_bkgd.clone_df, radius=50, chunk_size = 100) scipy.sparse.save_npz(os.path.join(project_path, f"{antigen_enriched_file}.rw_beta.npz"), tr.rw_beta) # Tip: For larger dataset you can use a sparse implementation: # 30.8 s ± 0 ns per loop ; tr.cpus = 6 # %timeit -r tr.compute_sparse_rect_distances(df = tr.clone_df, df2 = tr_bkdg.clone_df,radius=50, chunk_size=85) ############################################################################ # Step 5: Examine Density ECDFS ##### ############################################################################ # Investigate the density of neighbors to each TCR, based on expanding # distance radius. from tcrdist.ecdf import distance_ecdf, _plot_manuscript_ecdfs import matplotlib.pyplot as plt # Compute empirical cumulative density function (ecdf) # Compare Antigen Enriched TCRs (against itself). thresholds, antigen_enriched_ecdf = distance_ecdf( tr.pw_beta, thresholds=range(0,50,2)) # Compute empirical cumulative density function (ecdf) # Compare Antigen Enriched TCRs (against) 200K probability # inverse weighted background thresholds, background_ecdf = distance_ecdf( tr.rw_beta, thresholds=range(0,50,2), weights= tr_bkgd.clone_df['weights'], absolute_weight = True) # plot_ecdf similar to tcrdist3 manuscript # antigen_enriched_ecdf[antigen_enriched_ecdf == antigen_enriched_ecdf.min()] = 1E-10 f1 = _plot_manuscript_ecdfs( thresholds, antigen_enriched_ecdf, ylab= 'Proportion of Antigen Enriched TCRs', cdr3_len=tr.clone_df.cdr3_b_aa.str.len(), min_freq=1E-10) f1.savefig(os.path.join(project_path, f'{antigen_enriched_file}.ecdf_AER_plot.png')) f2 = _plot_manuscript_ecdfs( thresholds, background_ecdf, ylab= 'Proportion of Reference TCRs', cdr3_len=tr.clone_df.cdr3_b_aa.str.len(), min_freq=1E-10) f2.savefig(os.path.join(project_path, f'{antigen_enriched_file}.ecdf_BUR_plot.png')) ############################################################################ # Step 6: Find optimal radii (theta = 1E5 ##### ############################################################################ # To ascertain which meta-clonotypes are likely to be most specific, # take advantage of an existing function <bkgd_cntrl_nn2>. # d888 .d8888b. 8888888888 888888888 # d8888 d88P Y88b 888 888 # 888 888 888 888 888 # 888 888 888 8888888 8888888b. # 888 888 888 888 "Y88b # 888 888 888 888 888888 888 # 888 Y88b d88P 888 Y88b d88P # 8888888 "Y8888P" 8888888888 "Y8888P" level_tag = '1E5' from tcrdist.neighbors import bkgd_cntl_nn2 centers_df = bkgd_cntl_nn2( tr = tr, tr_background = tr_bkgd, weights = tr_bkgd.clone_df.weights, ctrl_bkgd = 10**-5, col = 'cdr3_b_aa', add_cols = ['v_b_gene', 'j_b_gene'], ncpus = 4, include_seq_info = True, thresholds = [x for x in range(0,50,2)], generate_regex = True, test_regex = True, forced_max_radius = 36) ############################################################################ # Step 6.2: (theta = 1E5) ALL meta-clonotypes .tsv file ## ############################################################################ # save center to project_path for future use centers_df.to_csv( os.path.join(project_path, f'{antigen_enriched_file}.centers_bkgd_ctlr_{level_tag}.tsv'), sep = "\t" ) # Many of meta-clonotypes contain redundant information. # We can winnow down to less-redundant list. We do this # by ranking clonotypes from most to least specific. # <min_nsubject> is minimum publicity of the meta-clonotype, # <min_nr> is minimum non-redundancy # Add neighbors, K_neighbors, and nsubject columns from tcrdist.public import _neighbors_variable_radius, _neighbors_sparse_variable_radius centers_df['neighbors'] = _neighbors_variable_radius(pwmat=tr.pw_beta, radius_list = centers_df['radius']) centers_df['K_neighbors'] = centers_df['neighbors'].apply(lambda x : len(x)) # We determine how many <nsubjects> are in the set of neighbors centers_df['nsubject'] = centers_df['neighbors'].\ apply(lambda x: tr.clone_df['subject'].iloc[x].nunique()) centers_df.to_csv( os.path.join(project_path, f'{antigen_enriched_file}.centers_bkgd_ctlr_{level_tag}.tsv'), sep = "\t" ) from tcrdist.centers import rank_centers ranked_centers_df = rank_centers( centers_df = centers_df, rank_column = 'chi2joint', min_nsubject = 2, min_nr = 1) ############################################################################ # Step 6.3: (theta = 1E5) NR meta-clonotypes .tsv file ### ############################################################################ # Output, ready to search bulk data. ranked_centers_df.to_csv( os.path.join(project_path, f'{antigen_enriched_file}.ranked_centers_bkgd_ctlr_{level_tag}.tsv'), sep = "\t" ) ############################################################################ # Step 6.4: (theta = 1E5) Output Meta-Clonotypes HTML Summary ### ############################################################################ # Here we can make a svg logo for each NR meta-clonotype if ranked_centers_df.shape[0] > 0: from progress.bar import IncrementalBar from tcrdist.public import make_motif_logo cdr3_name = 'cdr3_b_aa' v_gene_name = 'v_b_gene' svgs = list() svgs_raw = list() bar = IncrementalBar('Processing', max = ranked_centers_df.shape[0]) for i,r in ranked_centers_df.iterrows(): bar.next() centroid = r[cdr3_name] v_gene = r[v_gene_name] svg, svg_raw = make_motif_logo( tcrsampler = ts, pwmat = tr.pw_beta, clone_df = tr.clone_df, centroid = centroid , v_gene = v_gene , radius = r['radius'], pwmat_str = 'pw_beta', cdr3_name = 'cdr3_b_aa', v_name = 'v_b_gene', gene_names = ['v_b_gene','j_b_gene']) svgs.append(svg) svgs_raw.append(svg_raw) bar.next();bar.finish() ranked_centers_df['svg'] = svgs ranked_centers_df['svg_raw'] = svgs_raw def shrink(s): return s.replace('height="100%"', 'height="20%"').replace('width="100%"', 'width="20%"') labels =['cdr3_b_aa','v_b_gene', 'j_b_gene', 'pgen', 'radius', 'regex','nsubject','K_neighbors', 'bkgd_hits_weighted','chi2dist','chi2re','chi2joint'] output_html_name = os.path.join(project_path, f'{antigen_enriched_file}.ranked_centers_bkgd_ctlr_{level_tag}.html') # 888 888 88888888888 888b d888 888 # 888 888 888 8888b d8888 888 # 888 888 888 88888b.d88888 888 # 8888888888 888 888Y88888P888 888 # 888 888 888 888 Y888P 888 888 # 888 888 888 888 Y8P 888 888 # 888 888 888 888 " 888 888 # 888 888 888 888 888 88888888 with open(output_html_name, 'w') as output_handle: for i,r in ranked_centers_df.iterrows(): #import pdb; pdb.set_trace() svg, svg_raw = r['svg'],r['svg_raw'] output_handle.write("<br></br>") output_handle.write(shrink(svg)) output_handle.write(shrink(svg_raw)) output_handle.write("<br></br>") output_handle.write(pd.DataFrame(r[labels]).transpose().to_html()) output_handle.write("<br></br>") # To ascertain which meta-clonotypes are likely to be most specific, # take advantage of an existing function <bkgd_cntrl_nn2>. # d888 .d8888b. 8888888888 .d8888b. # d8888 d88P Y88b 888 d88P Y88b # 888 888 888 888 888 # 888 888 888 8888888 888d888b. # 888 888 888 888 888P "Y88b # 888 888 888 888 888888 888 888 # 888 Y88b d88P 888 Y88b d88P # 8888888 "Y8888P" 8888888888 "Y8888P" ############################################################################ # Step 6.5: Find optimal radii (theta = 1E6) ### ############################################################################ level_tag = '1E6' from tcrdist.neighbors import bkgd_cntl_nn2 centers_df = bkgd_cntl_nn2( tr = tr, tr_background = tr_bkgd, weights = tr_bkgd.clone_df.weights, ctrl_bkgd = 10**-6, col = 'cdr3_b_aa', add_cols = ['v_b_gene', 'j_b_gene'], ncpus = 4, include_seq_info = True, thresholds = [x for x in range(0,50,2)], generate_regex = True, test_regex = True, forced_max_radius = 36) ############################################################################ # Step 6.6: (theta = 1E6) ALL meta-clonotypes .tsv file ## ############################################################################ # save center to project_path for future use centers_df.to_csv( os.path.join(project_path, f'{antigen_enriched_file}.centers_bkgd_ctlr_{level_tag}.tsv'), sep = "\t" ) # Many of meta-clonotypes contain redundant information. # We can winnow down to less-redundant list. We do this # by ranking clonotypes from most to least specific. # <min_nsubject> is minimum publicity of the meta-clonotype, # <min_nr> is minimum non-redundancy # Add neighbors, K_neighbors, and nsubject columns from tcrdist.public import _neighbors_variable_radius, _neighbors_sparse_variable_radius centers_df['neighbors'] = _neighbors_variable_radius(pwmat=tr.pw_beta, radius_list = centers_df['radius']) centers_df['K_neighbors'] = centers_df['neighbors'].apply(lambda x : len(x)) # We determine how many <nsubjects> are in the set of neighbors centers_df['nsubject'] = centers_df['neighbors'].\ apply(lambda x: tr.clone_df['subject'].iloc[x].nunique()) centers_df.to_csv( os.path.join(project_path, f'{antigen_enriched_file}.centers_bkgd_ctlr_{level_tag}.tsv'), sep = "\t" ) from tcrdist.centers import rank_centers ranked_centers_df = rank_centers( centers_df = centers_df, rank_column = 'chi2joint', min_nsubject = 2, min_nr = 1) ############################################################################ # Step 6.7: (theta = 1E6) NR meta-clonotypes .tsv file ### ############################################################################ # Output, ready to search bulk data. ranked_centers_df.to_csv( os.path.join(project_path, f'{antigen_enriched_file}.ranked_centers_bkgd_ctlr_{level_tag}.tsv'), sep = "\t" ) ############################################################################ # Step 6.8: (theta = 1E6) Output Meta-Clonotypes HTML Summary ### ############################################################################ # Here we can make a svg logo for each meta-clonotype from progress.bar import IncrementalBar from tcrdist.public import make_motif_logo if ranked_centers_df.shape[0] > 0: cdr3_name = 'cdr3_b_aa' v_gene_name = 'v_b_gene' svgs = list() svgs_raw = list() bar = IncrementalBar('Processing', max = ranked_centers_df.shape[0]) for i,r in ranked_centers_df.iterrows(): bar.next() centroid = r[cdr3_name] v_gene = r[v_gene_name] svg, svg_raw = make_motif_logo( tcrsampler = ts, pwmat = tr.pw_beta, clone_df = tr.clone_df, centroid = centroid , v_gene = v_gene , radius = r['radius'], pwmat_str = 'pw_beta', cdr3_name = 'cdr3_b_aa', v_name = 'v_b_gene', gene_names = ['v_b_gene','j_b_gene']) svgs.append(svg) svgs_raw.append(svg_raw) bar.next();bar.finish() ranked_centers_df['svg'] = svgs ranked_centers_df['svg_raw'] = svgs_raw def shrink(s): return s.replace('height="100%"', 'height="20%"').replace('width="100%"', 'width="20%"') labels =['cdr3_b_aa', 'v_b_gene', 'j_b_gene', 'pgen', 'radius', 'regex','nsubject','K_neighbors', 'bkgd_hits_weighted','chi2dist','chi2re','chi2joint'] output_html_name = os.path.join(project_path, f'{antigen_enriched_file}.ranked_centers_bkgd_ctlr_{level_tag}.html') # 888 888 88888888888 888b d888 888 # 888 888 888 8888b d8888 888 # 888 888 888 88888b.d88888 888 # 8888888888 888 888Y88888P888 888 # 888 888 888 888 Y888P 888 888 # 888 888 888 888 Y8P 888 888 # 888 888 888 888 " 888 888 # 888 888 888 888 888 88888888 with open(output_html_name, 'w') as output_handle: for i,r in ranked_centers_df.iterrows(): #import pdb; pdb.set_trace() svg, svg_raw = r['svg'],r['svg_raw'] output_handle.write("<br></br>") output_handle.write(shrink(svg)) output_handle.write(shrink(svg_raw)) output_handle.write("<br></br>") output_handle.write(pd.DataFrame(r[labels]).transpose().to_html()) output_handle.write("<br></br>") if __name__ == "__main__": # 888 888 888 # 888 888 888 # 888 888 888 # 88888b. 888 8888b. .d8888b .d88b. 888888 .d8888b # 888 "88b 888 "88b 88K d8P Y8b 888 88K # 888 888 888 .d888888 "Y8888b. 88888888 888 "Y8888b. # 888 888 888 888 888 X88 Y8b. Y88b. X88 # 888 888 888 "Y888888 88888P' "Y8888 "Y888 88888P' # December 2020 # Seattle, WA # If run as a script, this will find meta-clonotypes for the 18 # SARS-CoV-2 MIRA sets with the most prior evidence of restriction # by as specific HLA-alleles. from collections import defaultdict import os import pandas as pd import re # <path> where files reside path = os.path.join(path_to_base,'tcrdist','data','covid19') # <all_files> list of all files all_files = [f for f in os.listdir(path) if f.endswith('.tcrdist3.csv')] # <restriction> list of tuples from Supporting Table S5 restriction = \ [('m_55_524_ALR','A*01'), ('m_1_8260_HTT','A*01'), ('m_45_689_SYF','A*01'), ('m_10_2274_LSP','B*07'), ('m_155_59_RAR','B*07'), ('m_133_102_NQK','B*15'), ('m_48_610_YLQ','A*02'), ('m_111_146_AEI','A*11'), ('m_53_532_NYL','A*24'), ('m_90_216_GYQ','C*07'), ('m_140_92_NSS','A*01'), ('m_55_524_ALR','B*08'), ('m_183_39_RIR','A*03'), ('m_10_2274_LSP','C*07'), ('m_99_191_QEC','B*40'), ('m_155_59_RAR','C*07'), ('m_185_39_ASQ','B*15'), ('m_147_73_DLF','B*08'), ('m_110_148_ELI','B*44'), ('m_51_546_AYK','A*03'), ('m_44_697_FPP','B*35'), ('m_118_136_ALN','A*11'), ('m_176_44_SST','A*11'), ('m_30_1064_KAY','B*57'), ('m_192_31_FQP','B*15'), ('m_70_345_DTD','A*01')] # <restrictions_dict> convert list to dictionary restrictions_dict = defaultdict(list) for k,v in restriction: restrictions_dict[k].append(v) # Loop through all files to construct a Dataframe of only those with strongest evidence of HLA-restriction cache = list() for f in all_files: rgs = re.search(pattern ='(mira_epitope)_([0-9]{1,3})_([0-9]{1,6})_([A-Z]{1,3})', string = f).groups() rgs4 = re.search(pattern ='(mira_epitope)_([0-9]{1,3})_([0-9]{1,6})_([A-Z]{1,4})', string = f).groups() key3 = f'm_{rgs[1]}_{rgs[2]}_{rgs[3]}' key4 = f'm_{rgs4[1]}_{rgs4[2]}_{rgs4[3]}' setkey = f"M_{rgs[1]}" include = key3 in restrictions_dict.keys() alleles = restrictions_dict.get(key3) cache.append((setkey, key3, key4, f, int(rgs[1]), int(rgs[2]), include, alleles)) # <hla_df> hla_df = pd.DataFrame(cache, columns = ['set', 'key3','key4','filename','set_rank','clones','hla_restricted','alleles']).\ sort_values(['hla_restricted','clones'], ascending = True).\ query('hla_restricted == True').reset_index(drop = True) from tcrdist.paths import path_to_base for ind,row in hla_df.iterrows(): file = row['filename'] print(file, ind) print(row) find_metaclonotypes(project_path = "hla_restricted_meta_clonotypes", source_path = os.path.join(path_to_base,'tcrdist','data','covid19'), antigen_enriched_file = file, ncpus = 6, seed = 3434) # 8888888888 888b 888 8888888b. # 888 8888b 888 888 "Y88b # 888 88888b 888 888 888 # 8888888 888Y88b 888 888 888 # 888 888 Y88b888 888 888 # 888 888 Y88888 888 888 # 888 888 Y8888 888 .d88P # 8888888888 888 Y888 8888888P" |
For each run, the function will write the following outputs:
project_folder/
├── .centers_bkgd_ctlr_1E5.tsv
├── .centers_bkgd_ctlr_1E6.tsv
├── .ecdf_AER_plot.png
├── .ecdf_BUR_plot.png
├── .olga100K_brit100K_bkgd.csv
├── .ranked_centers_bkgd_ctlr_1E5.html
├── .ranked_centers_bkgd_ctlr_1E5.tsv
├── .ranked_centers_bkgd_ctlr_1E6.html
├── .ranked_centers_bkgd_ctlr_1E6.tsv
└── .rw_beta.npz
One of the outputs is an html report of non-redundant meta-clonotypes .ranked_centers_bkgd_ctlr_1E6.html.
Meta-Clonotype Tabulation¶
The .ranked_centers_bkgd_ctlr_1E6.tsv files produced above can be used to directly search for the presence of meta-clonotypes is bulk data. How do we do this? Before showing how to do this for many files consider doing it for a single bulk repertoire:
In this example:
- We download a raw ImmunoSEQ file.
- Format it for use in tcrdist3.
- Search it with one of the meta-clonotypes file we made from above.
import os
import pandas as pd
from tcrdist.adpt_funcs import import_adaptive_file
from tcrdist.repertoire import TCRrep
from tcrdist.tabulate import tabulate
import math
import time
"""
You can download the file with wget or follow the link
!wget https://www.dropbox.com/s/1zbcf1ep8kgmlzy/KHBR20-00107_TCRB.tsv
"""
bulk_file = 'KHBR20-00107_TCRB.tsv'
df_bulk = import_adaptive_file(bulk_file)
df_bulk = df_bulk[['cdr3_b_aa',
'v_b_gene',
'j_b_gene',
'templates',
'productive_frequency',
'valid_cdr3']].\
rename(columns = {'templates':'count'})
df_bulk = df_bulk[(df_bulk['v_b_gene'].notna()) & (df_bulk['j_b_gene'].notna())].reset_index()
tr_bulk = TCRrep(cell_df = df_bulk,
organism = 'human',
chains = ['beta'],
db_file = 'alphabeta_gammadelta_db.tsv',
compute_distances = False)
search_file = os.path.join(
'tcrdist',
'data',
'covid19',
'mira_epitope_48_610_YLQPRTFL_YLQPRTFLL_YYVGYLQPRTF.tcrdist3.csv.ranked_centers_bkgd_ctlr_1E6.tsv')
df_search = pd.read_csv(search_file, sep = '\t')
df_search = df_search[['cdr3_b_aa','v_b_gene','j_b_gene','pgen','radius','regex']]
tr_search = TCRrep(cell_df = df_search,
organism = 'human',
chains = ['beta'],
db_file = 'alphabeta_gammadelta_db.tsv',
compute_distances = False)
tr_search.cpus = 1
tic = time.perf_counter()
tr_search.compute_sparse_rect_distances(df = tr_search.clone_df, df2 = tr_bulk.clone_df, chunk_size = 50, radius = 50)
results = tabulate(clone_df1 = tr_search.clone_df, clone_df2 = tr_bulk.clone_df, pwmat = tr_search.rw_beta)
toc = time.perf_counter()
print(f"TABULATED IN {toc - tic:0.4f} seconds")
The results, which can be saved as .tsv include the following:
cdr3_b_aa- meta-clonotype centroid CDR3v_b_gene- meta-clonotype centroid TRBVj_b_gene- meta-clonotype centroid TRBJpgen. - meta-clonotype centroid TRBV, CDR3 Pgen (Probability of Generation, estimated with OGLA)radius- meta-clonotype maximum TCRdist to find a neighborregex- pattern of conserved positions learned from the all those sequences with <radius> of the centroid in the antigen enriched dataset.cdr1_b_aa- meta-clonotype centroid CDR1cdr2_b_aa- meta-clonotype centroid CDR2pmhc_b_aa- meta-clonotype centroid CDR2.5bulk_sum_freq- In the bulk sample, sum of frequencies of TCRs within the <radius> of the centroid (* RADIUS)bulk_sum_counts- In the bulk sample, total template TCRs (counts) within the <radius> of the centroid (* RADIUS)bulk_seqs- In the bulk sample, CDR3 seqs of TCRs within the <radius> of the centroidbulk_v_genes- In the bulk sample, TRBVs of TCRs within the <radius> of the centroidbulk_j_genes- In the bulk sample, TRBJs of TCRs within the <radius> of the centroidbulk_distances- In the bulk sample, distances of TCRs from centroid, for those within the <radius> of the centroidbulk_counts- In the bulk sample, individual counts of each TCR within radius of the centroidbulk_freqs- In the bulk sample, individual frequencies of each TCR within radius of the centroidbulk_regex_match- In the bulk sample, boolean for each CDR3 within the <radius> whether it matched the regex motif patternbulk_sum_freqs_regex_adj- In the bulk sample, sum of frequencies of TCRs within the <radius> of the centroid and matching regex (RADIUS + MOTIF)bulk_sum_counts_regex_adj- In the bulk sample, sum of counts of TCRs within the <radius> of the centroid and matching regex (RADIUS + MOTIF)bulk_sum_freqs_tcrdist0- In the bulk sample, sum of frequencies of TCRs within the <radius> = 0 of the centroid (EXACT)bulk_sum_counts_tcrdist0- In the bulk sample, sum of counts of TCRs within the <radius> = 0 of the centroid (EXACT)
Tabulation Against Many¶
Below is a complete example used to tabulate the frequency and counts of each meta-clonotype in 694 bulk samples. Note that you have to supply a valid path to the directory where all the bulk files reside in your environment.
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 73 74 75 76 77 78 79 80 81 82 83 84 85 86 87 88 89 90 91 92 93 94 95 96 97 98 99 100 101 102 103 104 105 106 107 108 109 110 111 112 113 114 115 116 117 118 119 120 121 122 123 124 125 126 127 128 129 130 131 132 133 134 135 136 137 138 139 140 141 142 143 144 145 146 147 148 149 150 151 152 153 154 155 156 157 158 159 160 161 162 163 164 165 166 167 168 169 170 171 172 173 174 175 176 177 178 179 180 181 182 183 184 | # December 2020 # Seattle, WA import pandas as pd from tcrdist.repertoire import TCRrep from tcrdist.tabulate import tabulate import os import numpy as np from tcrdist.paths import path_to_base import time import math import parmap def get_safe_chunk(search_clones, bulk_clones,target = 10**7): """ This function help pick a chunk size that prevents excessive memory use, With two CPU, 10*7 should keep total overall memory demand below 1GB """ ideal_divisor = (search_clones * bulk_clones) / target if ideal_divisor < 1: ideal_chunk_size = search_clones print(ideal_chunk_size) else: ideal_chunk_size = math.ceil((search_clones)/ ideal_divisor) print(ideal_chunk_size) return ideal_chunk_size def do_search2(file, df_search, dest, tag): sample_name = file.replace('.tsv.tcrdist3.v_max.tsv','') tic = time.perf_counter() # <tr_search> tcrdist.repertoire.TCRrep object for computing distances tr_search = TCRrep(cell_df = df_search, organism = 'human', chains = ['beta'], db_file = 'alphabeta_gammadelta_db.tsv', compute_distances = False) # set cpus according to parameter above tr_search.cpus = 1 df_bulk = pd.read_csv(os.path.join(path, file), sep = '\t') df_bulk = df_bulk[['cdr3_b_aa','v_b_gene','j_b_gene','templates','productive_frequency']].rename(columns = {'templates':'count'}) tr_bulk = TCRrep( cell_df = df_bulk, organism = 'human', chains = ['beta'], db_file = 'alphabeta_gammadelta_db.tsv', compute_distances = False) #lines_per_file.append(tr_bulk.clone_df.shape[0]) search_clones = tr_search.clone_df.shape[0] bulk_clones = tr_bulk.clone_df.shape[0] # To avoid memory pressure on the system we set a target that tcrdist doesn't do more than 10M comparisons per process ideal_chunk_size = get_safe_chunk(tr_search.clone_df.shape[0], tr_bulk.clone_df.shape[0],target = 10**8) tr_search.compute_sparse_rect_distances(df = tr_search.clone_df, df2 = tr_bulk.clone_df, chunk_size = ideal_chunk_size) #(5) r1 = tabulate(clone_df1 = tr_search.clone_df, clone_df2 = tr_bulk.clone_df, pwmat = tr_search.rw_beta) outfile = os.path.join(dest, f"{sample_name}.{tag}.bulk_tabulation.tsv") print(f"WRITING: {outfile}") r1.to_csv(outfile, sep = '\t') toc = time.perf_counter() print(f"TABULATED IN {toc - tic:0.4f} seconds") del(tr_search) del(tr_bulk) return(f"{toc - tic:0.4f}s") if __name__ == "__main__": # 888 888 888 # 888 888 888 # 888 888 888 # 88888b. 888 8888b. .d8888b .d88b. 888888 .d8888b # 888 "88b 888 "88b 88K d8P Y8b 888 88K # 888 888 888 .d888888 "Y8888b. 88888888 888 "Y8888b. # 888 888 888 888 888 X88 Y8b. Y88b. X88 # 888 888 888 "Y888888 88888P' "Y8888 "Y888 88888P' # December 18, 2020 # Seattle, WA from collections import defaultdict import os import pandas as pd import re # <path> path where bulk files can be found path_bulkfiles = INSERT_HERE # <-------------------------- # <path> where files reside path = os.path.join(path_to_base,'tcrdist','data','covid19')#os.path.join('data-raw','2020-08-31-mira_tcr_by_epitope/') # <all_files> list of all files all_files = [f for f in os.listdir(path) if f.endswith('.tcrdist3.csv')] # <restriction> list of tuples from Supporting Table S5: https://docs.google.com/spreadsheets/d/1WAmze6lir-v11odO-nYYbCiYVh8WhQh_vy2d1UPPKb0/edit#gid=942295061 restriction = \ [('m_55_524_ALR','A*01'), ('m_1_8260_HTT','A*01'), ('m_45_689_SYF','A*01'), ('m_10_2274_LSP','B*07'), ('m_155_59_RAR','B*07'), ('m_133_102_NQK','B*15'), ('m_48_610_YLQ','A*02'), ('m_111_146_AEI','A*11'), ('m_53_532_NYL','A*24'), ('m_90_216_GYQ','C*07'), ('m_140_92_NSS','A*01'), ('m_55_524_ALR','B*08'), ('m_183_39_RIR','A*03'), ('m_10_2274_LSP','C*07'), ('m_99_191_QEC','B*40'), ('m_155_59_RAR','C*07'), ('m_185_39_ASQ','B*15'), ('m_147_73_DLF','B*08'), ('m_110_148_ELI','B*44'), ('m_51_546_AYK','A*03'), ('m_44_697_FPP','B*35'), ('m_118_136_ALN','A*11'), ('m_176_44_SST','A*11'), ('m_30_1064_KAY','B*57'), ('m_192_31_FQP','B*15'), ('m_70_345_DTD','A*01')] # <restrictions_dict> convert list to dictionary restrictions_dict = defaultdict(list) for k,v in restriction: restrictions_dict[k].append(v) # Loop through all files to construct a Dataframe of only those with strongest evidence of HLA-restriction cache = list() for f in all_files: rgs = re.search(pattern ='(mira_epitope)_([0-9]{1,3})_([0-9]{1,6})_([A-Z]{1,3})', string = f).groups() rgs4 = re.search(pattern ='(mira_epitope)_([0-9]{1,3})_([0-9]{1,6})_([A-Z]{1,4})', string = f).groups() key3 = f'm_{rgs[1]}_{rgs[2]}_{rgs[3]}' key4 = f'm_{rgs4[1]}_{rgs4[2]}_{rgs4[3]}' setkey = f"M_{rgs[1]}" include = key3 in restrictions_dict.keys() alleles = restrictions_dict.get(key3) cache.append((setkey, key3, key4, f, int(rgs[1]), int(rgs[2]), include, alleles)) # <hla_df> hla_df = pd.DataFrame(cache, columns = ['set', 'key3','key4','filename','set_rank','clones','hla_restricted','alleles']).\ sort_values(['hla_restricted','clones'], ascending = True).\ query('hla_restricted == True').reset_index(drop = True) for ind, row in hla_df.iterrows(): print(ind) print(row) tag_level = '1E6' # <tag> the results with this symbol tag = f"{row['set']}_{tag_level}" ranked_centers_fn = f"{row['filename']}.ranked_centers_bkgd_ctlr_{tag_level}.tsv" benchmark_fn = f"{row['filename']}.ranked_centers_bkgd_ctlr_{tag_level}.tsv.benchmark_tabulation.tsv" # <dest> place where .tsv files shall be saved dest = f'/fh/fast/gilbert_p/fg_data/tcrdist/t3/{tag}' if not os.path.isdir(dest): os.mkdir(dest) # <path> path where bulk files can be found path = path_bulkfiles # <covid_smaples> list of all samples to consider> covid_samples = ["BS-EQ-09-T1-replacement_TCRB", "BS-EQ-0022-T0-replacement_TCRB", "860011133_TCRB", "BS-GIGI_86-replacement_TCRB", "BS-EQ-12-T1-replacement_TCRB", "BS-GIGI_44-replacement_TCRB", "KH20-09697_TCRB", "BS-GN-0007-T0-replacement_TCRB", "860011216_TCRB", "BS-EQ-0002-T1-replacement_TCRB", "KH20-09988_TCRB", "BS-GIGI_21-replacement_TCRB", "INCOV018-AC-5_TCRB", "860011205_TCRB", "BS-GN-06-T0-replacement_TCRB", "KHBR20-00107_TCRB", "550043156_TCRB", "550041451_TCRB", "KH20-09700_TCRB", "BS-EQ-25-T0-replacement_TCRB", "860011320_TCRB", "BS-GN-0008-T0-replacement_TCRB", "BS-EQ-0002-T2-replacement_TCRB", "860011277_TCRB", "KH20-11306_TCRB", "BS-EQ-10-T1-replacement_TCRB", "BS-EQ-18-T1-replacement_TCRB", "KH20-11638_TCRB", "860011493_TCRB", "BS-GN-0010-T0-replacement_TCRB", "860011220_TCRB", "KH20-09681_TCRB", "550041430_TCRB", "INCOV033-AC-3_TCRB", "KHBR20-00093_TCRB", "860011221_TCRB", "BS-GIGI_28-replacement_TCRB", "BS-GIGI_46-replacement_TCRB", "KH20-09665_TCRB", "BS-EQ-0018-T0-replacement_TCRB", "860011287_TCRB", "BS-GIGI_42-replacement_TCRB", "INCOV004-BL-5_TCRB", "KH20-11809_TCRB", "BS-EQ-0001-T1-replacement_TCRB", "KH20-09671_TCRB", "KHBR20-00111_TCRB", "BS-EQ-0027-T1-replacement_TCRB", "BS-EQ-0024-T0-replacement_TCRB", "BS-GN-13-T0-replacement_TCRB", "BS-EQ-11-T2-replacement_TCRB", "BS-GIGI_32-replacement_TCRB", "BS-EQ-31-T0-replacement_TCRB", "BS-EQ-0027-T2-replacement_TCRB", "BS-GIGI_06-replacement_TCRB", "KH20-09679_TCRB", "KH20-09676_TCRB", "860011462_TCRB", "BS-GIGI_56-replacement_TCRB", "KH20-09722_TCRB", "KH20-09751_TCRB", "550040014_TCRB", "BS-GIGI_96-replacement_TCRB", "BS-GN-0012-T0-replacement_TCRB", "KHBR20-00085_TCRB", "KH20-09667_TCRB", "INCOV086-AC-3_TCRB", "KH20-11307_TCRB", "BS-GIGI_49-replacement_TCRB", "INCOV020-AC-5_TCRB", "BS-GIGI_85-replacement_TCRB", "KH20-11650_TCRB", "BS-GIGI_15-replacement_TCRB", "BS-EQ-43-T0-replacement_TCRB", "BS-GIGI_50-replacement_TCRB", "INCOV013-AC-5_TCRB", "BS-EQ-0011-T1-replacement_TCRB", "BS-GIGI_66-replacement_TCRB", "860011140_TCRB", "BS-EQ-12-T2-replacement_TCRB", "KHBR20-00089_TCRB", "860011235_TCRB", "860011123_TCRB", "KHBR20-00121_TCRB", "KHBR20-00180_TCRB", "BS-GIGI_18-replacement_TCRB", "BS-GIGI_64-replacement_TCRB", "BS-EQ-0024-T1-replacement_TCRB", "550042607_TCRB", "KH20-11683_TCRB", "KHBR20-00179_TCRB", "BS-GN-0002-T0-replacement_TCRB", "550043906_TCRB", "BS-EQ-0021-T0-replacement_TCRB", "BS-EQ-07-T1-replacement_TCRB", "860011306_TCRB", "550043943_TCRB", "BS-GIGI_20-replacement_TCRB", "BS-GIGI_52-replacement_TCRB", "KH20-09970_TCRB", "860011129_TCRB", "KHBR20-00098_TCRB", "860011124_TCRB", "860011110_TCRB", "BS-GIGI_01-replacement_TCRB", "KH20-09670_TCRB", "BS-GIGI_72-replacement_TCRB", "BS-EQ-11-T3-replacement_TCRB", "860011120_TCRB", "INCOV008-BL-5_TCRB", "BS-GN-0004-T0-replacement_TCRB", "INCOV028-AC-3_TCRB", "KH20-11295_TCRB", "860011214_TCRB", "BS-GIGI_13-replacement_TCRB", "860011204_TCRB", "BS-GIGI_87-replacement_TCRB", "860011215_TCRB", "860011202_TCRB", "INCOV005-BL-5_TCRB", "BS-GIGI_10-replacement_TCRB", "860011489_TCRB", "550043905_TCRB", "BS-GN-01-T0-replacement_TCRB", "550042578_TCRB", "KHBR20-00118_TCRB", "KH20-09655_TCRB", "BS-EQ-0016-T1-replacement_TCRB", "KHBR20-00106_TCRB", "BS-GIGI_62-replacement_TCRB", "BS-EQ-37-T0-replacement_TCRB", "860011244_TCRB", "KHBR20-00157_TCRB", "BS-GIGI_80-replacement_TCRB", "KHBR20-00182_TCRB", "KHBR20-00206_TCRB", "860011318_TCRB", "KH20-11645_TCRB", "KH20-09673_TCRB", "550040140_TCRB", "BS-GIGI_36-replacement_TCRB", "KHBR20-00171_TCRB", "BS-GIGI_81-replacement_TCRB", "860011225_TCRB", "550043911_TCRB", "BS-GIGI_03-replacement_TCRB", "KH20-09698_TCRB", "BS-EQ-29-T0-replacement_TCRB", "860011239_TCRB", "KH20-11303_TCRB", "550041293_TCRB", "INCOV028-BL-3_TCRB", "INCOV004-AC-5_TCRB", "110047437_TCRB", "860011201_TCRB", "BS-EQ-23-T1-replacement_TCRB", "KHBR20-00119_TCRB", "KHBR20-00149_TCRB", "BS-EQ-0017-T0-replacement_TCRB", "KH20-11810_TCRB", "BS-GIGI_45-replacement_TCRB", "550042567_TCRB", "860011109_TCRB", "KH20-09752_TCRB", "KH20-09675_TCRB", "KH20-09987_TCRB", "BS-GIGI_41-replacement_TCRB", "BS-EQ-0008-T1-replacement_TCRB", "550042420_TCRB", "KH20-09747_TCRB", "KHBR20-00144_TCRB", "KH20-11675_TCRB", "INCOV031-BL-3_TCRB", "KH20-11697_TCRB", "KHBR20-00141_TCRB", "KH20-09948_TCRB", "INCOV036-AC-3_TCRB", "550041421_TCRB", "BS-EQ-0026-T0-replacement_TCRB", "BS-GIGI_08-replacement_TCRB", "BS-EQ-29-T1-replacement_TCRB", "KHBR20-00160_TCRB", "860011322_TCRB", "KHBR20-00165_TCRB", "860011327_TCRB", "KH20-09674_TCRB", "KH20-09661_TCRB", "BS-GIGI_16-replacement_TCRB", "BS-EQ-43-T1_BS-GIGI-137-replacement_TCRB", "860011117_TCRB", "INCOV033-BL-3_TCRB", "KHBR20-00148_TCRB", "BS-GIGI_79-replacement_TCRB", "550042515_TCRB", "550042526_TCRB", "INCOV003-BL-5_TCRB", "BS-EQ-35-T0-replacement_TCRB", "550042550_TCRB", "BS-GN-0011-T0-replacement_TCRB", "BS-EQ-07-T2-replacement_TCRB", "KHBR20-00103_TCRB", "INCOV030-AC-3_TCRB", "KH20-09997_TCRB", "BS-GIGI_61-replacement_TCRB", "KHBR20-00161_TCRB", "KH20-09996_TCRB", "INCOV030-BL-3_TCRB", "KHBR20-00156_TCRB", "KHBR20-00137_TCRB", "550041125_TCRB", "KHBR20-00130_TCRB", "860011498_TCRB", "KH20-11698_TCRB", "BS-EQ-17-T1b-replacement_TCRB", "KHBR20-00105_TCRB", "550043986_TCRB", "INCOV071-BL-3_TCRB", "KH20-11304_TCRB", "BS-EQ-0023-T0-replacement_TCRB", "KH20-11692_TCRB", "550042377_TCRB", "KH20-09720_TCRB", "BS-GIGI_35-replacement_TCRB", "KHBR20-00113_TCRB", "860011105_TCRB", "KH20-11294_TCRB", "KHBR20-00167_TCRB", "INCOV031-AC-3_TCRB", "KH20-09666_TCRB", "KHBR20-00081_TCRB", "BS-GIGI_70-replacement_TCRB", "BS-GIGI_55-replacement_TCRB", "INCOV009-BL-5_TCRB", "BS-EQ-34-T0-replacement_TCRB", "BS-GIGI_34-replacement_TCRB", "KHBR20-00185_TCRB", "BS-EQ-33-T0-replacement_TCRB", "BS-GIGI_19-replacement_TCRB", "KHBR20-00126_TCRB", "550042351_TCRB", "KH20-11647_TCRB", "BS-GIGI_74-replacement_TCRB", "860011229_TCRB", "KH20-11640_TCRB", "BS-GN-0005-T0-replacement_TCRB", "KHBR20-00083_TCRB", "KH20-09735_TCRB", "550042361_TCRB", "KH20-09963_TCRB", "KHBR20-00166_TCRB", "KH20-11695_TCRB", "KHBR20-00096_TCRB", "BS-GIGI_07-replacement_TCRB", "KH20-11811_TCRB", "INCOV069-BL-3_TCRB", "BS-GIGI_05-replacement_TCRB", "BS-GIGI_40-replacement_TCRB", "INCOV053-AC-3_TCRB", "860011116_TCRB", "INCOV016-BL-5_TCRB", "860011112_TCRB", "INCOV034-BL-3_TCRB", "INCOV032-BL-3_TCRB", "KH20-09664_TCRB", "550041314_TCRB", "860011206_TCRB", "550042640_TCRB", "INCOV017-BL-5_TCRB", "BS-EQ-10-T2-replacement_TCRB", "KHBR20-00172_TCRB", "KH20-11813_TCRB", "550043940_TCRB", "550041390_TCRB", "INCOV015-BL-5_TCRB", "KH20-09684_TCRB", "BS-EQ-36-T0-replacement_TCRB", "KH20-11637_TCRB", "BS-EQ-39-T0-replacement_TCRB", "BS-EQ-16-T2-replacement_TCRB", "KHBR20-00153_TCRB", "BS-GIGI_92-replacement_TCRB", "BS-GIGI_91-replacement_TCRB", "860011492_TCRB", "KHBR20-00117_TCRB", "550041499_TCRB", "KHBR20-00181_TCRB", "KHBR20-00173_TCRB", "KHBR20-00168_TCRB", "KHBR20-00123_TCRB", "KH20-09945_TCRB", "BS-GN-0014-T0-replacement_TCRB", "550043912_TCRB", "KH20-09955_TCRB", "KH20-11298_TCRB", "550043995_TCRB", "KHBR20-00170_TCRB", "INCOV070-BL-3_TCRB", "KH20-11305_TCRB", "KHBR20-00203_TCRB", "KHBR20-00188_TCRB", "BS-GIGI_53-replacement_TCRB", "INCOV036-BL-3_TCRB", "KH20-09959_TCRB", "KHBR20-00145_TCRB", "BS-EQ-33-T1-replacement_TCRB", "KH20-09677_TCRB", "INCOV087-BL-3_TCRB", "INCOV084-BL-3_TCRB", "KH20-11642_TCRB", "KH20-09687_TCRB", "KH20-09724_TCRB", "KH20-11687_TCRB", "BS-EQ-14-T3-replacement_TCRB", "KH20-11688_TCRB", "INCOV016-AC-5_TCRB", "BS-EQ-38-T1_BS-GIGI-117-replacement_TCRB", "INCOV075-AC-3_TCRB", "KHBR20-00162_TCRB", "550043973_TCRB", "550042259_TCRB", "550042183_TCRB", "BS-EQ-18-T2-replacement_TCRB", "KH20-11672_TCRB", "KH20-11643_TCRB", "KH20-11639_TCRB", "BS-GIGI_26-replacement_TCRB", "KH20-09696_TCRB", "KH20-09952_TCRB", "860011325_TCRB", "550043955_TCRB", "KHBR20-00177_TCRB", "BS-EQ-15-T1-replacement_TCRB", "BS-EQ-34-T1-replacement_TCRB", "860011312_TCRB", "KH20-11671_TCRB", "KHBR20-00196_TCRB", "KH20-09695_TCRB", "860011490_TCRB", "INCOV005-AC-5_TCRB", "550043980_TCRB", "KH20-09734_TCRB", "KH20-09750_TCRB", "KHBR20-00094_TCRB", "860011243_TCRB", "KHBR20-00143_TCRB", "BS-EQ-28-T1-replacement_TCRB", "KH20-09947_TCRB", "860011309_TCRB", "860011450_TCRB", "INCOV049-AC-3_TCRB", "860011310_TCRB", "860011113_TCRB", "KH20-09964_TCRB", "KH20-09991_TCRB", "KH20-11292_TCRB", "860011131_TCRB", "KHBR20-00200_TCRB", "KHBR20-00078_TCRB", "KHBR20-00186_TCRB", "BS-GIGI_27-replacement_TCRB", "550041250_TCRB", "KH20-09962_TCRB", "860011127_TCRB", "KH20-09999_TCRB", "860011338_TCRB", "KHBR20-00076_TCRB", "860011137_TCRB", "KHBR20-00204_TCRB", "550042523_TCRB", "INCOV017-AC-5_TCRB", "550043908_TCRB", "KHBR20-00205_TCRB", "KHBR20-00158_TCRB", "KH20-09708_TCRB", "860011218_TCRB", "860011382_TCRB", "BS-EQ-36-T1-replacement_TCRB", "860011125_TCRB", "BS-EQ-41-T1-replacement_TCRB", "BS-EQ-33-T2-replacement_TCRB", "860011132_TCRB", "550043971_TCRB", "860011246_TCRB", "INCOV006-BL-5_TCRB", "KHBR20-00134_TCRB", "550041349_TCRB", "KH20-10000_TCRB", "KH20-09718_TCRB", "KH20-09694_TCRB", "550042609_TCRB", "860011383_TCRB", "KH20-09741_TCRB", "550043927_TCRB", "KH20-11807_TCRB", "KH20-09668_TCRB", "KH20-11682_TCRB", "550042599_TCRB", "KHBR20-00127_TCRB", "550042631_TCRB", "KHBR20-00102_TCRB", "KH20-11814_TCRB", "KH20-09992_TCRB", "KHBR20-00178_TCRB", "KH20-09701_TCRB", "860011240_TCRB", "BS-EQ-41-T0-replacement_TCRB", "INCOV014-BL-5_TCRB", "KH20-11659_TCRB", "INCOV007-BL-5_TCRB", "KH20-09685_TCRB", "BS-EQ-28-T2-replacement_TCRB", "INCOV035-AC-3_TCRB", "INCOV011-AC-5_TCRB", "BS-GN-0016-T0-replacement_TCRB", "KHBR20-00152_TCRB", "BS-GN-0009-T0-replacement_TCRB", "INCOV025-AC-3_TCRB", "KHBR20-00128_TCRB", "550043167_TCRB", "INCOV051-AC-3_TCRB", "860011314_TCRB", "KHBR20-00108_TCRB", "KH20-09704_TCRB", "KH20-09749_TCRB", "KH20-09956_TCRB", "550042447_TCRB", "860011212_TCRB", "KH20-11670_TCRB", "BS-EQ-0028-T0-replacement_TCRB", "550040030_TCRB", "KH20-09746_TCRB", "KH20-11296_TCRB", "860011233_TCRB", "860011139_TCRB", "BS-GN-0003-T0-replacement_TCRB", "INCOV073-BL-3_TCRB", "860011203_TCRB", "KH20-09971_TCRB", "INCOV007-AC-5_TCRB", "KHBR20-00135_TCRB", "KH20-11291_TCRB", "860011241_TCRB", "KH20-09736_TCRB", "KHBR20-00201_TCRB", "BS-EQ-26-T1-replacement_TCRB", "KH20-11657_TCRB", "860011231_TCRB", "860011227_TCRB", "KHBR20-00082_TCRB", "KH20-09657_TCRB", "KH20-09966_TCRB", "KH20-09725_TCRB", "KH20-11681_TCRB", "550042442_TCRB", "KHBR20-00109_TCRB", "860011210_TCRB", "KH20-09985_TCRB", "860011238_TCRB", "550044028_TCRB", "860011445_TCRB", "550042400_TCRB", "BS-EQ-30-T2-replacement_TCRB", "KHBR20-00084_TCRB", "550042395_TCRB", "KHBR20-00136_TCRB", "KH20-11641_TCRB", "KH20-11302_TCRB", "KH20-11676_TCRB", "BS-EQ-0014-T1-replacement_TCRB", "KH20-11673_TCRB", "860011111_TCRB", "KH20-09967_TCRB", "KH20-09984_TCRB", "KH20-11300_TCRB", "KH20-11297_TCRB", "860011228_TCRB", "KH20-11301_TCRB", "KH20-11646_TCRB", "KHBR20-00151_TCRB", "KH20-11661_TCRB", "KH20-11299_TCRB", "KHBR20-00131_TCRB", "KH20-09709_TCRB", "860011496_TCRB", "KH20-09946_TCRB", "KHBR20-00163_TCRB", "550041393_TCRB", "KH20-09690_TCRB", "KHBR20-00080_TCRB", "KH20-09753_TCRB", "KH20-09669_TCRB", "KHBR20-00139_TCRB", "KH20-09691_TCRB", "INCOV019-AC-5_TCRB", "860011217_TCRB", "KH20-09980_TCRB", "INCOV034-AC-3_TCRB", "KH20-09951_TCRB", "BS-EQ-0003-T1-replacement_TCRB", "KH20-09978_TCRB", "KH20-11660_TCRB", "KH20-09961_TCRB", "INCOV035-BL-3_TCRB", "KH20-09990_TCRB", "860011303_TCRB", "KHBR20-00184_TCRB", "KH20-09954_TCRB", "INCOV044-AC-3_TCRB", "KHBR20-00154_TCRB", "KHBR20-00183_TCRB", "KHBR20-00122_TCRB", "KH20-09993_TCRB", "KH20-09748_TCRB", "550042547_TCRB", "KH20-09689_TCRB", "KH20-09727_TCRB", "BS-EQ-0014-T2-replacement_TCRB", "860011323_TCRB", "KH20-11812_TCRB", "KHBR20-00091_TCRB", "INCOV048-BL-3_TCRB", "KH20-09977_TCRB", "KH20-09986_TCRB", "KH20-09745_TCRB", "860011115_TCRB", "KHBR20-00087_TCRB", "550043914_TCRB", "550043981_TCRB", "860011499_TCRB", "INCOV054-BL-3_TCRB", "KHBR20-00100_TCRB", "550042239_TCRB", "KH20-11649_TCRB", "KH20-09953_TCRB", "860011304_TCRB", "KHBR20-00133_TCRB", "550042210_TCRB", "INCOV051-BL-3_TCRB", "860011497_TCRB", "KHBR20-00116_TCRB", "KH20-09715_TCRB", "BS-EQ-30-T1-replacement_TCRB", "860011348_TCRB", "860011118_TCRB", "KH20-09975_TCRB", "550043928_TCRB", "KH20-09714_TCRB", "860011242_TCRB", "KHBR20-00079_TCRB", "550041441_TCRB", "KH20-09960_TCRB", "860011326_TCRB", "KHBR20-00191_TCRB", "BS-GIGI_75-replacement_TCRB", "KH20-09968_TCRB", "KH20-09723_TCRB", "KH20-09979_TCRB", "KH20-11658_TCRB", "INCOV047-BL-3_TCRB", "INCOV002-AC-5_TCRB", "KH20-09731_TCRB", "INCOV029-AC-3_TCRB", "KH20-11674_TCRB", "110047542_TCRB", "KH20-09743_TCRB", "KH20-11678_TCRB", "KH20-11689_TCRB", "KH20-09699_TCRB", "KH20-09726_TCRB", "860011336_TCRB", "860011256_TCRB", "KHBR20-00097_TCRB", "INCOV006-AC-5_TCRB", "KHBR20-00086_TCRB", "KH20-11651_TCRB", "KHBR20-00125_TCRB", "KH20-09755_TCRB", "KH20-11655_TCRB", "KH20-09683_TCRB", "KH20-09976_TCRB", "860011280_TCRB", "INCOV047-AC-3_TCRB", "KH20-11684_TCRB", "860011219_TCRB", "KH20-09654_TCRB", "INCOV044-BL-3_TCRB", "KH20-09738_TCRB", "INCOV015-AC-5_TCRB", "INCOV050-AC-3_TCRB", "860011494_TCRB", "KH20-11662_TCRB", "KH20-11665_TCRB", "INCOV062-BL-3_TCRB", "860011226_TCRB", "KH20-09702_TCRB", "860011354_TCRB", "860011236_TCRB", "KHBR20-00095_TCRB", "860011209_TCRB", "860011317_TCRB", "KH20-09711_TCRB", "KH20-11699_TCRB", "KH20-09754_TCRB", "KHBR20-00190_TCRB", "860011388_TCRB", "860011356_TCRB", "550043920_TCRB", "KHBR20-00099_TCRB", "KHBR20-00129_TCRB", "KH20-11696_TCRB", "KH20-11686_TCRB", "KH20-11664_TCRB", "INCOV026-BL-3_TCRB", "KH20-11667_TCRB", "KH20-11666_TCRB", "860011347_TCRB", "KH20-11648_TCRB", "KH20-09739_TCRB", "550044000_TCRB", "KH20-09972_TCRB", "KH20-11668_TCRB", "860011138_TCRB", "860011208_TCRB", "550041239-1_TCRB", "550042542_TCRB", "860011223_TCRB", "KHBR20-00092_TCRB", "INCOV052-BL-3_TCRB", "INCOV050-BL-3_TCRB", "860011311_TCRB", "550044029_TCRB", "INCOV003-AC-5_TCRB", "KHBR20-00088_TCRB", "860011264_TCRB", "INCOV012-AC-5_TCRB", "INCOV001-AC-5_TCRB", "INCOV002-BL-5_TCRB", "KH20-11654_TCRB", "KHBR20-00115_TCRB", "KH20-09707_TCRB", "860011392_TCRB", "KH20-11656_TCRB", "KH20-09717_TCRB", "860011284_TCRB", "KHBR20-00174_TCRB", "KH20-11653_TCRB", "KH20-09719_TCRB", "KH20-09958_TCRB", "INCOV026-AC-3_TCRB", "860011119_TCRB", "KH20-09981_TCRB", "KH20-11652_TCRB", "860011250_TCRB", "KH20-09995_TCRB", "INCOV059-AC-3_TCRB", "860011282_TCRB", "KH20-11669_TCRB", "550041173_TCRB", "KH20-09973_TCRB", "KH20-09706_TCRB", "860011122_TCRB", "KH20-09969_TCRB", "KH20-11293_TCRB", "INCOV062-AC-3_TCRB", "860011340_TCRB", "INCOV055-BL-3_TCRB", "860011224_TCRB", "KH20-09658_TCRB", "KHBR20-00104_TCRB", "860011211_TCRB", "KH20-11679_TCRB", "860011130_TCRB", "860011495_TCRB", "INCOV027-BL-3_TCRB", "KH20-09693_TCRB", "KHBR20-00146_TCRB", "860011106_TCRB", "KH20-09721_TCRB", "INCOV077-BL-3_TCRB", "KH20-09716_TCRB", "INCOV027-AC-3_TCRB", "860011121_TCRB", "KH20-11644_TCRB", "INCOV045-BL-3_TCRB", "860011330_TCRB", "INCOV023-AC-3_TCRB", "860011260_TCRB", "KH20-09994_TCRB", "KH20-09744_TCRB", "KH20-09712_TCRB", "860011488_TCRB", "550042451_TCRB", "INCOV078-BL-3_TCRB", "KHBR20-00164_TCRB"] # <max number of cpus to use> ncpus = 2 # <df_search> dataframe containing metaclonotypes df_search = pd.read_csv(os.path.join('hla_restricted_meta_clonotypes', ranked_centers_fn), sep = "\t") if df_search.shape[0] > 0: df_search = df_search[['cdr3_b_aa','v_b_gene','j_b_gene','pgen','radius','regex']] # <all_files> list of all files in the project <path> all_files = [f for f in os.listdir(path) if f.endswith('v_max.tsv')] # add file size all_files = [ (f, f.replace('.tsv.tcrdist3.v_max.tsv',''), os.stat(os.path.join(path,f)).st_size) for f in all_files ] # sort ascending by file size, smaller files first all_files = sorted(all_files, key=lambda x: x[2]) # <all_files_ms> is <all_files> subset to only the samples with 30 days of COVID19 diagnosis, and not including ADPATIVE-COVID19 training samples all_files_ms = [(x,y,z) for x,y,z in all_files if y in covid_samples] ts_iterable = [f for f,sn,_ in all_files_ms] import parmap timings = parmap.map(do_search2, ts_iterable, df_search = df_search.copy(), dest = dest, tag = tag, pm_processes=6, pm_pbar = True) pd.DataFrame({'file':ts_iterable, 'seconds':timings}).to_csv(os.path.join('hla_restricted_meta_clonotypes', benchmark_fn), sep = "\t") # 8888888888 888b 888 8888888b. # 888 8888b 888 888 "Y88b # 888 88888b 888 888 888 # 8888888 888Y88b 888 888 888 # 888 888 Y88b888 888 888 # 888 888 Y88888 888 888 # 888 888 Y8888 888 .d88P # 8888888888 888 Y888 8888888P" |